Scope of traudem
What this package does
- It provides a guide to installation of TauDEM and its dependencies GDAL and MPI, depending on your operating system.
- It checks that TauDEM and its dependencies are correctly installed and included to the PATH.
- It provides wrapper commands for calling TauDEM methods from R.
What this package does not
- It does not provide an automatic installation of TauDEM and its
dependencies. Please refer to the
vignette("taudem-installation")for instructions on this. - It does not provide extensive documentation on the functioning of TauDEM commands. Please refer to the original TauDEM documentation for that.
Can traudem run all TauDEM methods?
Yes! Some TauDEM methods have dedicated wrappers like
taudem_pitremove(), with argument names that we strove to
make informative. For other methods, you can use
taudem_exec() and provide the arguments as TauDEM CLI would
expect them. You could make a PR to this repository to add more
dedicated wrappers.
Installation
Refer to vignette("taudem-installation", package = "traudem").
In particular, after installing the TauDEM CLI and dependencies as well
as the R package, please run traudem::taudem_sitrep().
traudem::taudem_sitrep()
#> ✔ Found GDAL version 3.0.4.
#> ✔ Found mpiexec (OpenRTE) 4.0.3 (MPI).
#> ✔ Found TauDEM path (/usr/local/taudem).
#> ✔ Found TauDEM executables directory (/usr/local/taudem).
#> ✔ Found all TauDEM executables.
#> ℹ Testing TauDEM on an example file (please wait a bit)...
#> ── TauDEM output ────────────────────────────────────────────────────────────────────
#> PitRemove version 5.3.9
#> Input file DEM.tif has projected coordinate system.
#> Nodata value input to create partition from file: nan
#> Nodata value recast to float used in partition raster: nan
#> Processes: 1
#> Header read time: 0.008202
#> Data read time: 0.003174
#> Compute time: 0.028233
#> Write time: 0.002115
#> Total time: 0.041723
#> This run may take on the order of 1 minutes to complete.
#> This estimate is very approximate.
#> Run time is highly uncertain as it depends on the complexity of the input data
#> and speed and memory of the computer. This estimate is based on our testing on
#> a dual quad core Dell Xeon E5405 2.0GHz PC with 16GB RAM.
#> ── End of TauDEM output ─────────────────────────────────────────────────────────────
#> ✔ Was able to launch a TauDEM example!
#> ! Double-check above output for serious error messages.You can install traudem from CRAN:
install.packages("traudem")You can install the development version of traudem from R-universe:
# Enable repository from lucarraro
options(
repos = c(
lucarraro = 'https://lucarraro.r-universe.dev',
CRAN = 'https://cloud.r-project.org'
)
)
# Download and install traudem in R
install.packages('traudem')Or from GitHub with:
# install.packages("devtools")
devtools::install_github("lucarraro/traudem")Simple example
This is how you would use traudem to launch TauDEM’s PitRemove:
library(traudem)
test_dir <- withr::local_tempdir()
fs::file_copy(
system.file("test-data", "DEM.tif", package = "traudem"),
file.path(test_dir, "DEM.tif")
)
output <- taudem_pitremove(file.path(test_dir, "DEM.tif"))
#> PitRemove version 5.3.9
#> Input file /tmp/Rtmp64HAv3/filefc9c38806e1b/DEM.tif has projected coordinate system.
#> Nodata value input to create partition from file: nan
#> Nodata value recast to float used in partition raster: nan
#> Processes: 1
#> Header read time: 0.002983
#> Data read time: 0.001157
#> Compute time: 0.016390
#> Write time: 0.001719
#> Total time: 0.022249
#> This run may take on the order of 1 minutes to complete.
#> This estimate is very approximate.
#> Run time is highly uncertain as it depends on the complexity of the input data
#> and speed and memory of the computer. This estimate is based on our testing on
#> a dual quad core Dell Xeon E5405 2.0GHz PC with 16GB RAM.
output
#> [1] "/tmp/Rtmp64HAv3/filefc9c38806e1b/DEMfel.tif"We ran the example above in a temporary directory that
withr automatically deletes. If you want to automatically
get rid of some of the intermediary files created by TauDEM in one of
your pipelines, you might be interested in
withr::local_tempfile().
If you wanted to run this same code without seeing the messages from
TauDEM, you can either use the quiet argument:
test_dir <- withr::local_tempdir()
fs::file_copy(
system.file("test-data", "DEM.tif", package = "traudem"),
file.path(test_dir, "DEM.tif")
)
output <- taudem_pitremove(file.path(test_dir, "DEM.tif"), quiet = TRUE)
output
#> [1] "/tmp/Rtmp64HAv3/filefc9c7b85bbf2/DEMfel.tif"Or set the traudem.quiet option
(options(traudem.quiet = TRUE) or for just the session,
withr::local_options(traudem.quiet = TRUE)):
withr::local_options(traudem.quiet = TRUE)
test_dir <- withr::local_tempdir()
fs::file_copy(
system.file("test-data", "DEM.tif", package = "traudem"),
file.path(test_dir, "DEM.tif")
)
output <- taudem_pitremove(file.path(test_dir, "DEM.tif"))
output
#> [1] "/tmp/Rtmp64HAv3/filefc9c2c87ac26/DEMfel.tif"Full example: download DEM data and extract a river network
traudem can be used in combination with R-package elevatr to extract
a river network from elevation data retrieved from the web.
The following example shows how to derive the contour of a catchment and drainage area values for all cells within that contour by using the D8 flow direction algorithm.
The only required inputs are:
- coordinates of the catchment outlet;
- coordinates of the lower-left and top-right corners of a rectangular area containing the catchment of interest;
- EPSG code corresponding to the projection used to express coordinate data (see also https://epsg.org/home.html);
- zoom level at which the DEM data are extracted (see
elevatrdocumentation).
library(elevatr)
library(sf)
#> Linking to GEOS 3.8.0, GDAL 3.0.4, PROJ 6.3.1; sf_use_s2() is TRUE
library(terra)
#> terra 1.6.17
library(traudem)
library(shapefiles)
#> Loading required package: foreign
#>
#> Attaching package: 'shapefiles'
#> The following objects are masked from 'package:foreign':
#>
#> read.dbf, write.dbf
library(withr)
taudem_sitrep() # verify that TauDEM is correctly installed
#> ✔ Found GDAL version 3.0.4.
#> ✔ Found mpiexec (OpenRTE) 4.0.3 (MPI).
#> ✔ Found TauDEM path (/usr/local/taudem).
#> ✔ Found TauDEM executables directory (/usr/local/taudem).
#> ✔ Found all TauDEM executables.
#> ℹ Testing TauDEM on an example file (please wait a bit)...
#> ── TauDEM output ────────────────────────────────────────────────────────────────────
#> PitRemove version 5.3.9
#> Input file DEM.tif has projected coordinate system.
#> Nodata value input to create partition from file: nan
#> Nodata value recast to float used in partition raster: nan
#> Processes: 1
#> Header read time: 0.002094
#> Data read time: 0.000788
#> Compute time: 0.013749
#> Write time: 0.001609
#> Total time: 0.018240
#> This run may take on the order of 1 minutes to complete.
#> This estimate is very approximate.
#> Run time is highly uncertain as it depends on the complexity of the input data
#> and speed and memory of the computer. This estimate is based on our testing on
#> a dual quad core Dell Xeon E5405 2.0GHz PC with 16GB RAM.
#> ── End of TauDEM output ─────────────────────────────────────────────────────────────
#> ✔ Was able to launch a TauDEM example!
#> ! Double-check above output for serious error messages.
test_dir <- local_tempdir() # temporary directory storing intermediary files created by TauDEM
# Input data for the river Wigger (central Switzerland)
EPSG <- 21781 # EPSG code corresponding to CH1903 (Swiss projected coordinate system)
x_outlet <- 637478 # outlet x coordinate in CH1903 coordinates
y_outlet <- 237413 # outlet y coordinate in CH1903 coordinates
x_ll <- 620000 # x coordinate of the lower-left (SW) limit of the region
x_tr <- 660000 # x coordinate of the top-right (NE) limit of the region
y_ll <- 200000 # y coordinate of the lower-left (SW) limit of the region
y_tr <- 250000 # y coordinate of the top-right (NE) limit of the region
zoom <- 9 # corresponds to cellsize ~= 100 m at the latitude of interest
# use elevatr to download DEM
loc.df <- data.frame(x=c(x_ll, x_tr), y=c(y_ll,y_tr))
r <- rast()
crs(r) <- paste0("epsg:", EPSG)
crs_str <- crs(r)
z <- get_elev_raster(locations = loc.df, prj = crs_str, z = zoom, verbose = F)
#> Warning in showSRID(SRS_string, format = "PROJ", multiline = "NO", prefer_proj = prefer_proj): PROJ/GDAL PROJ string degradation in workflow
#> repeated warnings suppressed
#> Discarded datum CH1903 in Proj4 definition
#>
#> Accessing raster elevation [-------------------------] 0%
Accessing raster elevation [=====>-------------------] 25%
Accessing raster elevation [===========>-------------] 50%
Accessing raster elevation [==================>------] 75%
Accessing raster elevation [=========================] 100%
#> Mosaicing & Projecting
z <- rast(z)
z <- crop(z, ext(x_ll, x_tr, y_ll, y_tr))
# reclassify: all pixels with elev=NA are set to 0. Then the pit remove algorithm will take care of them
z <- classify(z, matrix(c(NA, NA, 0), 1, 3))
writeRaster(z, filename = file.path(test_dir, "DEM.tif")) # write elevation raster to temporary directory
# apply TauDEM functions
# Remove pits
out_fel <- taudem_pitremove(file.path(test_dir, "DEM.tif"), quiet = TRUE)
# D8 flow directions
out_p <- taudem_d8flowdir(out_fel, quiet = TRUE)
out_p <- out_p$output_d8flowdir_grid # file path of flow direction file
# Contributing area
out_ad8 <- taudem_aread8(out_p, quiet = TRUE)
# Threshold
out_src <- taudem_threshold(out_ad8, quiet = TRUE)
# create shapefile with approximate outlet location
shp.point <- function(x, y, sname = "shape"){ # function to create point shapefile given coordinates
n <- length(x)
dd <- data.frame(Id = 1:n, X = x, Y = y)
ddTable <- data.frame(Id = c(1), Name = paste("Outlet", 1:n, sep = ""))
ddShapefile <- convert.to.shapefile(dd, ddTable, "Id", 1)
write.shapefile(ddShapefile, sname, arcgis = T)
invisible(paste0(sname,".shp"))
}
out_shp <- shp.point(x_outlet, y_outlet, file.path(test_dir, "ApproxOutlet"))
# Move outlet to stream
out_moved.shp <- taudem_moveoutletstostream(out_p, out_src, outlet_file = out_shp,
output_moved_outlets_file = file.path(test_dir, "Outlet.shp"),
quiet = TRUE)
# Contributing area upstream of outlet
out_ssa <- taudem_aread8(out_p, outlet_file = out_moved.shp, quiet = TRUE)
# Derive catchment contour
ssa <- rast(out_ssa)
# reclassify: pixels within the catchment have value 1, others 0
ssa_cont <- classify(ssa, matrix(c(NA, NA, 0, 1, Inf, 1), 2, 3, byrow = T))
# plot DEM of the region and catchment contours
plot(z, col = terrain.colors(1000), range = c(0,1200))
contour(ssa_cont, add = T, labels = "")
title("DEM and catchment contour")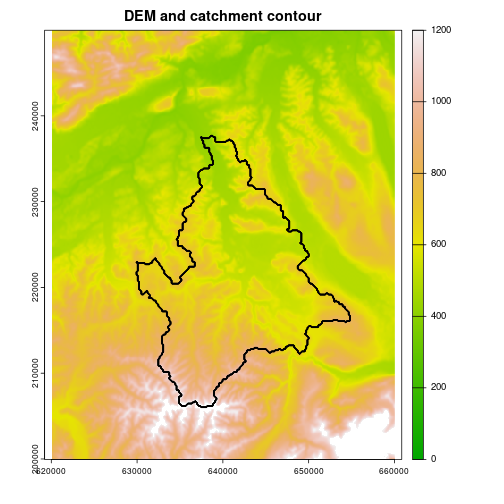
# plot map of drainage area within the catchment
plot(ssa, col = hcl.colors(1000, "viridis"))
title("Drainage area [no. cells]")
# total catchment area
ssa_log <- ssa
values(ssa_log) <- log10(values(ssa))
plot(ssa_log, col = hcl.colors(1000, "viridis"))
title("Drainage area [log10(no. cells)]")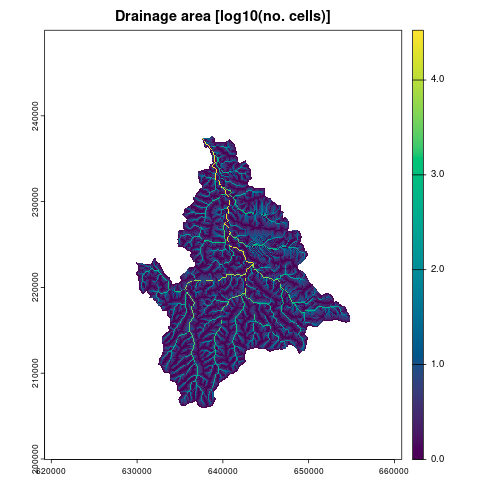
cellsize <- sqrt(prod(res(z))) # cellsize is geometric mean of downloaded raster resolution
max(values(ssa),na.rm=T)*cellsize^2 # total catchment area in m^2
#> [1] 363822337The total area of the extracted catchment is 364 km2.
What should I do if I do not retrieve the desired catchment?
It might happen that, even when the coordinates of the catchment
outlet are precisely attributed, the obtained catchment shape is not the
desired one (in particular, much smaller than the sought one). This
might be due to issues in determining the correct outlet position in the
extracted river network. In fact, the outlet position is moved downslope
from the initial, user-specified location
(x_outlet, y_outlet) to the closest stream cell via
function taudem_moveoutletstostream. Depending on the
terrain topography and the DEM resolution, the extracted river network
might not overlap exactly with real river networks, hence the possible
errors.
To solve this issue, it is suggested to:
- Slightly change the coordinates
x_outlet,y_outlet, to better align the input outlet location with the extracted stream cells. - Change argument
threshold_parameterintaudem_threshold. Stream cells are defined as cells with drainage area not smaller thanthreshold_parameter(expressed in number of cells). Ifthreshold_parameteris too large, the actual outlet location will be moved too far away in the downstream direction; if too small, the actual outlet location will not be moved enough in the downstream direction.